GENOTYPIC CHARACTERIZATION OF PSEUDOMONAS AERUGINOSA FROM POINT-OF-SALES DEVICES IN LOKOJA, NIGERIA
DOI:
https://doi.org/10.33003/fjs-2025-0911-3737Keywords:
Radiation, Radio-frequency, Smartphones, Spectrum Analyzer, Human healthAbstract
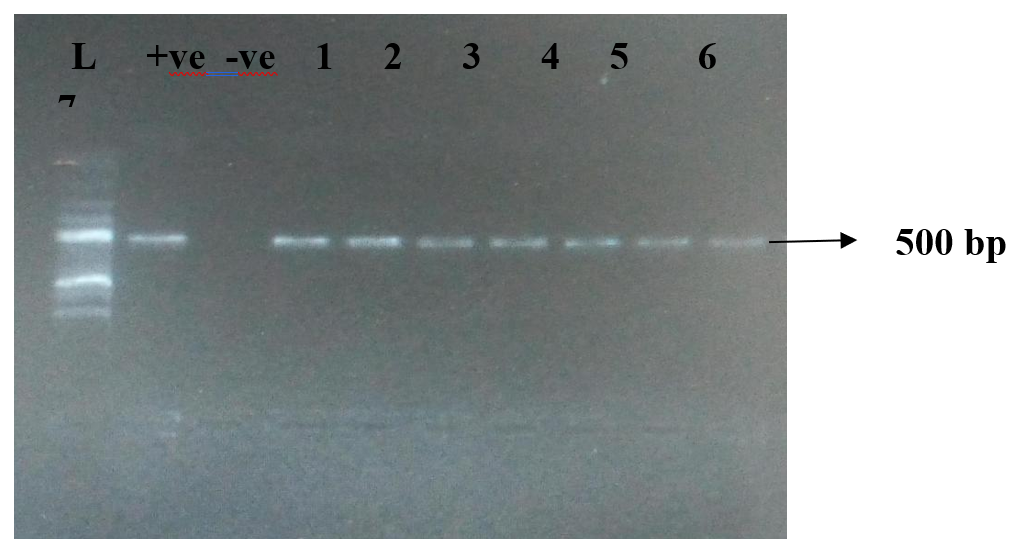
The presence of Pseudomonas aeruginosa on shared frequently used surfaces, such as Point-of-Sales (POS) devices present serious public health problem. This study assessed P. aeruginosa on POS devices frequently used within Lokoja metropolis, with the aim of determining the presence of antibiotic resistance and virulence genes in the P. aeruginosa. Following standard procedures, Seventy-two POS devices were randomly sampled using sterile swabs soaked in peptone water and cultured on sterile agar plates. Biochemical characterization of isolates was carried out while antibiotics susceptibility testing was carried out using the Kirby-Bauer disc diffusion method. Some resistance and virulence genes of P. aeruginosa were detected using polymerase chain reaction (PCR) assay. Bacterial isolates were characterized as Staphylococcus aureus (32.4%), Klebsiella pneumoniae (21.9%), Pseudomonas aeruginosa (17.2%), Escherichia coli (14.2%), Bacillus subtilis (10.1%), Enterobacter spp. (2.4 %) and Enterococcus sp. (1.8%). Pseudomonas aeruginosa isolates displayed antibiotic resistance against tetracyclines (58.6%), followed by ceftazidime (51.7%), gentamicin (48.3%), cefepime (48.3%), ceftriaxone (48.3%), cefoxitin (44.8%), levofloxacin (41.2%), ciprofloxacin (37.9%), amoxicillin-clavulanic acid (37.9%) and imipenem (24.1%). For the virulence genes, 72.4%, 38%, 28% of the Pseudomonas aeruginosa strains were positive for OprL, toxA and lasB, respectively while for the antibiotic resistance genes, 14% were positive for blaVIM and 10.3% for blaIMP. This study reveals the presence of pathogenic P. aeruginosa on POS devices with their virulence genes and antibiotics resistance genes. This emphasizes the health threat associated with shared surfaces such as the POS devices. Thus, users must maintain sanitary and hygienic practices while using these devices.
References
REFERENCES
Adenipekun, E. O., Akinleye, E. F., Tewogbade, O. A., and Iwalokun, B. A. (2023). Detection of virulence genes and multidrug resistance in Pseudomonas aeruginosa clinical isolates from a public hospital in Lagos, Nigeria. Scientific African, 22, e01950.
Aduku, A. E., Bakare, A. A., Oyabambi, A., Akanni, E. O., and Isah, F. I. (2025). IMPACT OF POINT OF SALE (POS) TERMINAL ADOPTION ON THE RETURN ON PROFIT AFTER TAX (PAT) OF TIER-ONE BANKS IN NIGERIA. Prestieesci Research Review, 2(1), 710–735.
Aghamiri, S., Amirmozafari, N., Fallah Mehrabadi, J., Fouladtan, B., and Samadi Kafil, H. (2014). Antibiotic Resistance Pattern and Evaluation of Metallo‐Beta Lactamase Genes Including bla‐IMP and bla‐VIM Types in Pseudomonas aeruginosa Isolated from Patients in Tehran Hospitals. International Scholarly Research Notices, 2014(1), 941507.
Assefa, M., and Amare, A. (2022). Biofilm-associated multi-drug resistance in hospital-acquired infections: A review. Infection and Drug Resistance, 5061–5068.
Bernal-Rosas, Y., Osorio-Muñoz, K., and Torres-García, O. (2015). Pseudomonas aeruginosa: An emerging nosocomial trouble in veterinary. Revista MVZ Córdoba, 20, 4937–4946.
Brzozowski, M., Krukowska, Ż., Galant, K., Jursa-Kulesza, J., and Kosik-Bogacka, D. (2020). Genotypic characterisation and antimicrobial resistance of Pseudomonas aeruginosa strains isolated from patients of different hospitals and medical centres in Poland. BMC Infectious Diseases, 20, 1–9.
Chand, Y., Khadka, S., Sapkota, S., Sharma, S., Khanal, S., Thapa, A., Rayamajhee, B., Khadka, D. K., Panta, O. P., and Shrestha, D. (2021). Clinical Specimens are the Pool of oprL and toxA Virulence Genes Harboring Multidrug-Resistant Pseudomonas aeruginosa: Findings from a Tertiary Hospital of Nepal. Emergency Medicine International.
Clinical and Laboratory Standards Institute (CLSI) (2018) Performance Standards for Antimicrobial Susceptibility Testing. CLSI Approved Standard M100-S15. Clinical and Laboratory Standards Institute, Wayne.
Dawodu, O., and Akanbi, R. (2021). Isolation and identification of microorganisms associated with automated teller machines on Federal Polytechnic Ede campus. PLoS One, 16(8), e0254658.
Douraghi, M., Ghasemi, F., Dallal, M. S., Rahbar, M., and Rahimiforoushani, A. (2014). Molecular identification of Pseudomonas aeruginosa recovered from cystic fibrosis patients. Journal of Preventive Medicine and Hygiene, 55(2), 50.
Farhan, R. E., Solyman, S. M., Hanora, A. M., and Azab, M. M. (2023). Molecular detection of different virulence factors genes harbor pslA, pelA, exoS, toxA and algD among biofilm-forming clinical isolates of Pseudomonas aeruginosa. Cellular and Molecular Biology, 69(5), 32–39.
Fernández-Billón, M., Llambías-Cabot, A. E., Jordana-Lluch, E., Oliver, A., and Macià, M. D. (2023). Mechanisms of antibiotic resistance in Pseudomonas aeruginosa biofilms. Biofilm, 5, 100129.
Gauba, A., and Rahman, K. M. (2023). Evaluation of antibiotic resistance mechanisms in gram-negative bacteria. Antibiotics, 12(11), 1590.
Harb, C. P. (2017). Genome-wide antibiotic resistance and virulence profiling of Pseudomonas Aeruginosa isolated from clinical samples in Lebanon.(c2017).
Horcajada, J. P., Montero, M., Oliver, A., Sorlí, L., Luque, S., Gómez-Zorrilla, S., Benito, N., and Grau, S. (2019). Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clinical Microbiology Reviews, 32(4), 10–1128.
Innocent-Adiele, H., Nkara, N., Osuala, O., Daokoru-Olukole, C., Okafor, O., Onu, E., Cookey, T., Nwankwo, G., Mbah, E., and Adim, C. (2023). Isolation and Antibiogram Pattern of Bacteria Isolated from Automated Teller Machine (ATM) at Abuja Campus, University of Port Harcourt, Nigeria. Int. J. Pathog. Res, 12(6), 127–135.
Jafari-Sales, A., and Khaneshpour, H. (2020). Molecular Study of BlaIMP and BlaVIM Genes in Pseudomonas Aeruginosa Strains, Producer of Metallo Beta Lactamases Isolated from Clinical Samples in Hospitals and Medical Centers of Tabriz. Paramedical Sciences and Military Health, 14(4), 18–25.
Kaye, K. S., and Pogue, J. M. (2015). Infections caused by resistant gram‐negative bacteria: Epidemiology and management. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy, 35(10), 949–962.
Khan, A., Miller, W. R., and Arias, C. A. (2018). Mechanisms of antimicrobial resistance among hospital-associated pathogens. Expert Review of Anti-Infective Therapy, 16(4), 269–287.
Lepe, J. A., and Martínez-Martínez, L. (2022). Resistance mechanisms in Gram-negative bacteria. Medicina Intensiva (English Edition), 46(7), 392–402.
Li, X.-Z., Plésiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clinical Microbiology Reviews, 28(2), 337–418.
Mamudu, Z. U., and Gayovwi, G. O. (2019). Cashless policy and its impact on the Nigerian economy. International Journal of Education and Research, 7(3), 111–132.
Manner, C., Dias Teixeira, R., Saha, D., Kaczmarczyk, A., Zemp, R., Wyss, F., Jaeger, T., Laventie, B.-J., Boyer, S., and Malone, J. G. (2023). A genetic switch controls Pseudomonas aeruginosa surface colonization. Nature Microbiology, 8(8), 1520–1533.
Mendes, R. E., Kiyota, K. A., Monteiro, J., Castanheira, M., Andrade, S. S., Gales, A. C., Pignatari, A. C., and Tufik, S. (2007). Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. Journal of Clinical Microbiology, 45(2), 544–547.
Mentasti, M., Prime, K., Sands, K., Khan, S., and Wootton, M. (2019). Rapid detection of IMP, NDM, VIM, KPC and OXA-48-like carbapenemases from Enterobacteriales and Gram-negative non-fermenter bacteria by real-time PCR and melt-curve analysis. European Journal of Clinical Microbiology and Infectious Diseases, 38, 2029–2036.
Moradali, M. F., Ghods, S., and Rehm, B. H. (2017). Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Frontiers in Cellular and Infection Microbiology, 7, 39.
Mostafa, A. E., Hassan, M. G., El-Metwally, M. M., Abo Neima, S. E., and Zewail, M. (2024). Bacteria Associated with Automated Teller Machines: Isolation and Identification. Egyptian Academic Journal of Biological Sciences, G. Microbiology, 16(2), 139–149.
Ntanda, P. M. (2024). Clinical and molecular evaluation of pseudomonas aeruginosa nosocomial infections among adult patients at the university teaching hospital in Lusaka province and Ndola teaching hospital in Copperbelt province of Zambia.
Odewale, G., Jibola-Shittu, M.Y., Bala, H.E.M., Akogwu, R., Raimi, L.O. (2024). Antibiotic susceptibility and detection of extended-spectrum β-lactamase genes in gram-negative bacteria isolated from automated teller machines. FUDMA Journal of Sciences, 8(1):118-124. DOI: https://doi.org/10.33003/fjs-2024-0801-2207.
Porter, L., Sultan, O., Mitchell, B. G., Jenney, A., Kiernan, M., Brewster, D. J., and Russo, P. L. (2024). How long do nosocomial pathogens persist on inanimate surfaces? A scoping review. Journal of Hospital Infection, 147, 25–31.
Qin, S., Xiao, W., Zhou, C., Pu, Q., Deng, X., Lan, L., Liang, H., Song, X., and Wu, M. (2022). Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduction and Targeted Therapy, 7(1), 199.
Queenan, A. M., and Bush, K. (2007). Carbapenemases: The versatile β-lactamases. Clinical Microbiology Reviews, 20(3), 440–458.
Reynolds, D., and Kollef, M. (2021). The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: An update. Drugs, 81(18), 2117–2131.
Rizvi, S., Kurtz, A., Williams, I., Gualdoni, J., Myzyri, I., and Wheeler, M. (2020). Protecting financial transactions through networks and point of sales. Journal of Cyber Security Technology, 4(4), 211–239.
Rocha, A. J., Barsottini, M. R. de O., Rocha, R. R., Laurindo, M. V., Moraes, F. L. L. de, and Rocha, S. L. da. (2019). Pseudomonas aeruginosa: Virulence factors and antibiotic resistance genes. Brazilian Archives of Biology and Technology, 62, e19180503.
Rubin, J., Walker, R., Blickenstaff, K., Bodeis-Jones, S., and Zhao, S. (2008). Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomonas aeruginosa isolated from canine infections. Veterinary Microbiology, 131(1–2), 164–172.
Urban-Chmiel, R., Marek, A., Stępień-Pyśniak, D., Wieczorek, K., Dec, M., Nowaczek, A., and Osek, J. (2022). Antibiotic resistance in bacteria—A review. Antibiotics, 11(8), 1079.
Uriawan, W., Faroj, R. Z., Hadid, R. A., Khoirunnisa, S., Julianto, S., and Sopian, A. R. (2024). Innovative Data Management Strategies in Point of Sale Application Development: Increasing Business Productivity.
Wu, Y. T., Zhu, L. S., Tam, K. C., Evans, D. J., and Fleiszig, S. M. (2015). Pseudomonas aeruginosa survival at posterior contact lens surfaces after daily wear. Optometry and Vision Science, 92(6), 659–664.
Yang, J., Xu, J.-F., and Liang, S. (2024). Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and emerging treatment. Critical Reviews in Microbiology, 1–19.
Yusha’u, M., Ahmad, J., Adam, R., and Hamza, M. (2024). Investigating the contamination levels, bacterial diversity and susceptibility pattern of bacteria isolated from point of sale (pos) devices in Kano metropolis. Bayero Journal of Pure and Applied Sciences, 17(1), 43–50.
Zafer, M. M., Mohamed, G. A., Ibrahim, S. R., Ghosh, S., Bornman, C., and Elfaky, M. A. (2024). Biofilm-mediated infections by multidrug-resistant microbes: A comprehensive exploration and forward perspectives. Archives of Microbiology, 206(3), 101.
Downloads
Published
Issue
Section
Categories
License
Copyright (c) 2025 Motunrayo Yemisi Jibola-Shittu, Gbolabo Odewale, Mela Ilu Luka, Joshua Atabo Odoh, Hannatu Eleojo Mary Bala, Aminat Bukola Kehinde

This work is licensed under a Creative Commons Attribution 4.0 International License.




